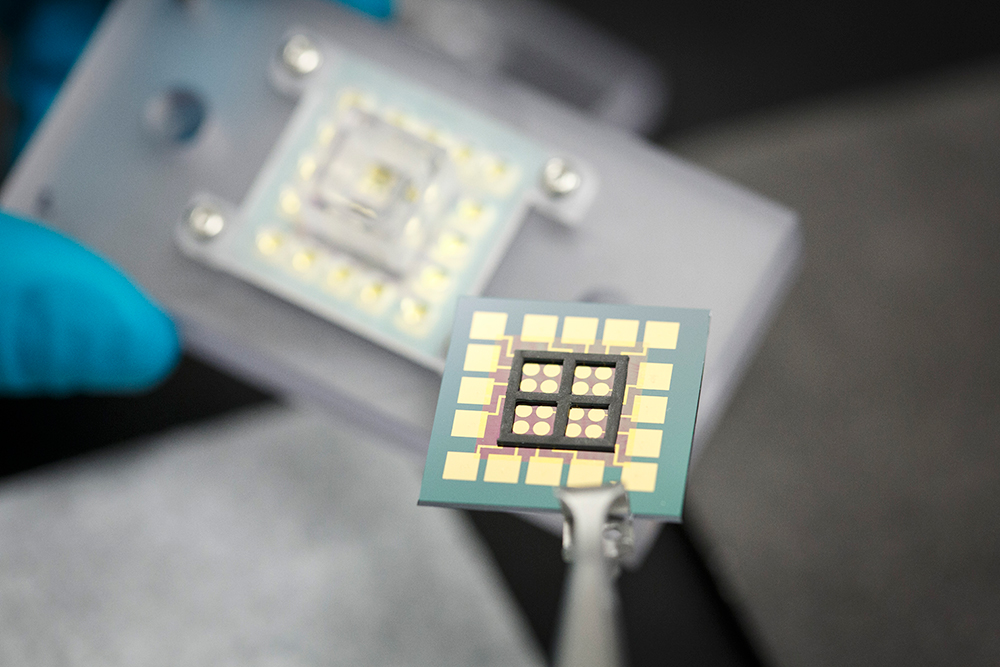
Multiple DNA strands are anchored to gold electrodes inside the black square areas on this chip’s surface. The chip connects to other components in a device UT Dallas researchers designed to mimic aspects of a human cell. This allows them to study the effects of anti-cancer drugs on simulated cancer cells.
A UT Dallas physicist has developed a novel technology that not only sheds light on basic cell biology, but also could aid in the development of more effective cancer treatments or early diagnosis of disease.
Dr. Jason Slinker, associate professor of physics, and his colleagues developed an electronic device that uses DNA molecules — the genetic material found in every human cell — and other biochemicals to simulate certain cell activity.
They started with an electronic “chip,” studded with strands of DNA, that Slinker designed, and combined it with other proteins and enzymes to mimic features of a cellular environment.
“We can’t replicate the entire cell, but our DNA chip-based platform simulates a nucleus, which contains a cell’s DNA, along with the nuclear membrane and the cytoplasm with a few cell components added,” Slinker said. “We combine these in a way that lets us study complex biochemical processes, including how cells repair DNA that has been damaged.”
It’s well-known that the biochemistry of cancer cells or other diseased cells differs from that of healthy cells, and that those differences can affect the way cells respond to drugs. For example, the concentrations of various proteins and enzymes can vary widely between cancer and normal cells. Some anti-cancer drugs take advantage of these differences, targeting and killing cancer cells while leaving most normal cells alone. Such drugs often have fewer side effects in patients.
“With our DNA chip technology, we have control over which biological factors we include in our simulated cell,” Slinker said. “We can change the composition to compare the differences in key biochemical reactions in a healthy cell and a cancer cell, for example. We also can test the effects of various drugs on these biochemical pathways, with multiple tests on the same chip. We think this could be an important tool to develop more effective treatments for patients.”
At the core of the cell-simulation device is the DNA chip that Slinker designed. His research combines biology with physics, with the aim of incorporating DNA into nanoscale electronics and biosensors.

Dr. Jason Slinker
Utilizing the UT Dallas Cleanroom Research Laboratory, Slinker manufactured the chip to exploit the fact that DNA molecules — shaped like lengths of twisted ladders — can carry electric charge, similar to the way electricity flows through a wire. Multiple DNA strands are anchored to gold electrodes on the chip’s surface, resulting in what looks like a forest of tiny nanoscale trees.
When a DNA molecule is damaged, a “break” occurs in the strand, which alters or eliminates the electrical signal flowing through each strand. The DNA chip registers those changes in the electrical signal. By monitoring that immediate electrical output, researchers can track and characterize in real time the DNA damage and the activity of proteins involved in making repairs.
“This technology gives us insight not only into the underlying biology of DNA damage and repair, but it can also tell us more precisely what happens when an anti-cancer drug interacts with certain cellular proteins and enzymes,” Slinker said.
In a recent study, Slinker’s research group tested whether their simulated cell could track the activity of beta-lapachone, an anti-cancer drug. The drug works by homing in on NQO1, an enzyme that is elevated in a number of cancers, but is at relatively low levels in normal cells. When the drug interacts with NQO1 in cancer cells, it initiates a chain of biochemical reactions that damages the cells’ DNA.
But that damage is not what directly kills the cancer cells.
“Beta-lapachone makes the cell’s DNA repair mechanism go into overdrive,” said Dr. David A. Boothman, the Robert B. and Virginia Payne Endowed Professor of Oncology, and professor of oncology, pharmacology and radiation oncology at UT Southwestern Medical Center in Dallas and a collaborator on Slinker’s research. He is the associate director for translational research at UT Southwestern’s Simmons Comprehensive Cancer Center, and has been researching beta-lapachone and its effects on cancer cells for more than 15 years.
“The drug causes tumor-selective DNA damage dependent on NQO1 expression, then the natural enzymes in the cell go crazy. Cells lose all of the energy they carry and die,” Boothman said.
“This type of device could eventually develop into a fast and inexpensive diagnostic tool. Or it might be used to further personalized medicine, where multiple drugs could be tested on a small patient blood sample to determine in advance of treatment which drugs will be most effective for that particular patient.”
Dr. Jason Slinker,
associate professor of physics
in the School of Natural Sciences and Mathematics
With Boothman’s input, Slinker’s group made a simulated cell with elevated NQO1 levels and other enzymes assumed to be involved in beta-lapachone-induced cell death. In a set of proof-of-concept experiments, they showed that their DNA chip could successfully track DNA repair activity in simulated cells that have been treated with beta-lapachone. They describe their efforts in a study published recently in the journal Biosensors and Bioelectronics.
Slinker said there are a number of potential applications of the technology to public health, and moving forward, his group will be investigating those possibilities. For example, a small blood sample from a patient might reveal an elevated level of DNA-repair activity, which can be an early indicator of cancer or other disease states.
“This type of device could eventually develop into a fast and inexpensive diagnostic tool,” he said. “Or it might be used to further personalized medicine, where multiple drugs could be tested on a small patient blood sample to determine in advance of treatment which drugs will be most effective for that particular patient.”
Other UT Dallas researchers involved in the work are lead study author and physics doctoral student Dimithree Kahanda MS’13, and former Slinker lab member Dr. Marc McWilliams BS’08, MS’10, PhD’15. In addition to Boothman, UT Southwestern medical/graduate student Dr. Gaurab Chakrabarti also participated.
The study was supported by UT Dallas faculty startup funds, a gift from Plexon Inc., and the National Cancer Institute, part of the National Institutes of Health.